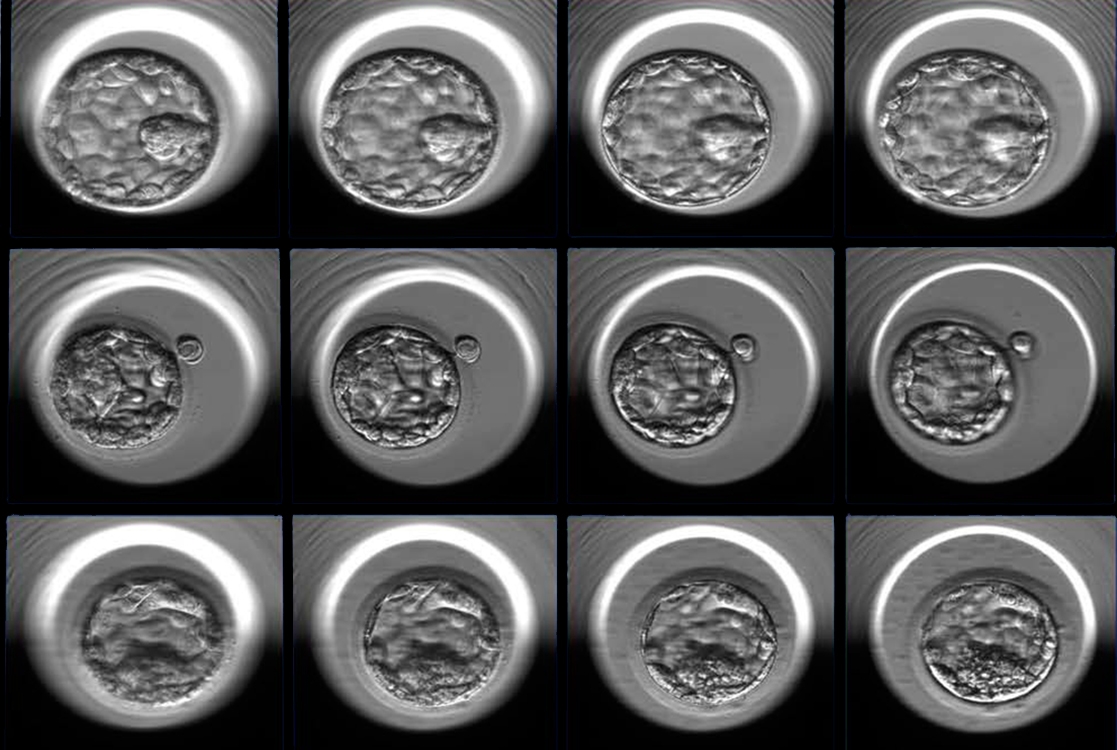
A new artificial intelligence approach by Weill Cornell Medicine investigators can identify with a great degree of accuracy whether a 5-day-old, in vitro fertilized human embryo has a high potential to progress to a successful pregnancy. The technique, which analyzes time-lapse images of the early-stage embryos, could reduce the number of in vitro fertilization cycles it takes to achieve a successful pregnancy, improve the success rate of IVF and minimize the risk of multiple pregnancies.
Infertility is estimated to affect about 8 percent of women of child-bearing age. While IVF has helped millions give birth, the average success rate in the United States is approximately 45 percent.
For the study, published April 4 in NPJ Digital Medicine, investigators used 12,000 photos of human embryos taken precisely 110 hours after fertilization to train an artificial intelligence algorithm to discriminate between poor and good embryo quality. To arrive at this designation, each embryo was first assigned a grade by embryologists that considered various aspects of the embryo’s appearance. The investigators then performed a statistical analysis to correlate the embryo grade with the probability of going on to a successful pregnancy outcome. Embryos were considered good quality if the chances were greater than 58 percent and poor quality if the chances were below 35 percent. After training and validation, the algorithm, dubbed Stork, was able to classify the quality of a new set of images with 97 percent accuracy.
“By introducing new technology into the field of IVF we can automate and standardize a process that was very dependent on subjective human judgement. This pioneering work gives us a window into how this field might look in the future,” said Dr. Zev Rosenwaks, director and physician-in-chief of the Ronald O. Perelman and Claudia Cohen Center for Reproductive Medicine at NewYork-Presbyterian/Weill Cornell Medical Center and Weill Cornell Medicine, and the Revlon Distinguished Professor of Reproductive Medicine in Obstetrics and Gynecology at Weill Cornell Medicine.
Choosing the embryo with the best chances of developing into a healthy pregnancy is currently a subjective process. Agreement is low among even experienced embryologists as to how to predict the viability of an individual embryo based upon its appearance at the blastocyst stage, in which it consists of only 200-300 cells.
“We wanted to develop an objective method that can be used to standardize and optimize the selection process to increase the success rates of IVF,” said Dr. Nikica Zaninovic, co-senior author and director of the Embryology Lab in the Center for Reproductive Medicine.
In a collaboration between the Center for Reproductive Medicine and the Caryl and Israel Englander Institute for Precision Medicine at Weill Cornell Medicine, the investigators spent more than six months reviewing approximately 50,000 anonymized images, representing 10,148 human embryos, collected by time-lapse photography over seven years. With the embryologist-assigned grade and the hindsight knowledge of the pregnancy outcome, the investigators could classify the embryos as good, fair or poor quality. Ultimately, they used two sets of 6,000 images, good or poor quality, to teach the algorithm how to classify new images presented to it.
“This is the first time, to our knowledge, that anyone has applied a deep learning algorithm on human embryos with such a large number of images,” said Dr. Pegah Khosravi, the lead author of the study and a postdoctoral associate in computational biomedicine.
Deep learning is an artificial intelligence approach that is roughly modeled after the neural networks of the brain, which analyze information in increasing layers of complexity. As the computer is fed new information, its ability to recognize the desired patterns, whether they are the features of a healthy embryo or the cells comprising a lung cancer tumor, improves automatically. The size of the training data set is critically important to the success of the algorithm, with more data leading to better outcomes.
“Our algorithm will help embryologists maximize the chances that their patients will have a single healthy pregnancy,” said Dr. Olivier Elemento, director of the Englander Institute for Precision Medicine and associate director of the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine at Weill Cornell Medicine. “The IVF procedure will remain the same, but we’ll be able to improve outcomes by harnessing the power of artificial intelligence.”
While Stork can select good quality embryos with a high degree of accuracy, previous studies have suggested that only 80 percent of the pregnancy success rate relies on the embryo quality. Maternal age, in particular, is associated with a decreasing rate of successful embryo implantation in the uterus.
Fertility specialists often implant multiple embryos to try to maximize the chances of having one successful birth, but the process is imprecise and can result in multiple pregnancies, which carries its own risks, such as low birth weight, premature delivery and maternal complications. Thus the investigators developed another computational approach that can take into account maternal age and the quality of multiple embryos to determine the best combination to achieve a single live birth.
“We are trying to tailor the process for the individual patient, because not every patient is the same,” said Dr. Zaninovic, who is also associate professor of embryology in clinical obstetrics and gynecology and in clinical reproductive medicine at Weill Cornell Medicine. “We want to do personalized medicine with precision medicine to get the best result.”
Using clinical data for 2,182 of the embryos, the investigators created a decision tree to assess the successful pregnancy rate by using a combination of embryo quality and patient age, as the most important clinical variable. They also provided a probability analysis aiming to optimize embryo selection and maximize likelihood of single pregnancy.
Stork is currently an investigative tool and the researchers plan to incorporate additional clinical and technical parameters to improve the algorithm.
“It’s very important that we could put a team together here that contains computer scientists, precision medicine experts, embryologists and clinicians,” said Dr. Iman Hajirasouliha, co-senior author and assistant professor of computational genomics in computational biomedicine in the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine, assistant professor of physiology and biophysics and a member of the Englander Institute for Precision Medicine. “We needed a strong team with a wide area of expertise to solve this problem.”