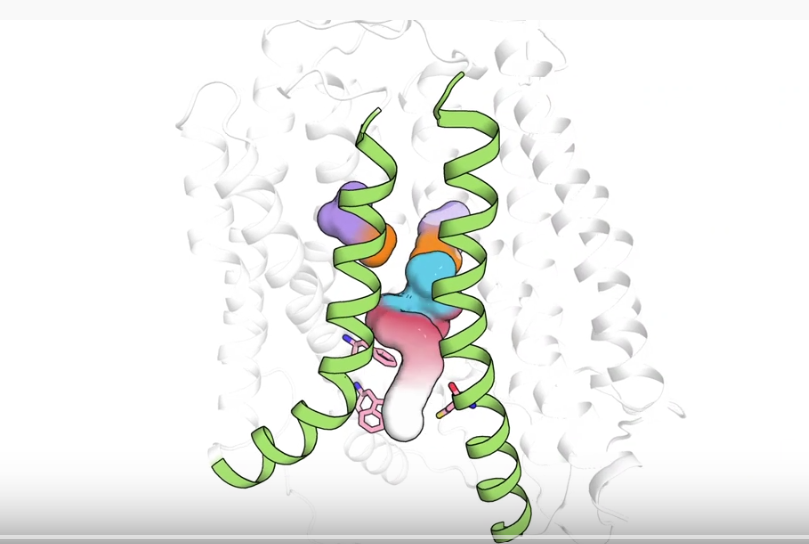
The structure and function of an important protein that transports essential omega-3 fatty acid molecules from the bloodstream into the brain has been determined by a team led by scientists at Columbia University Vagelos College of Physicians and Surgeons, Weill Cornell Medicine, and Duke-NUS Medical School. The findings illuminate a crucial aspect of brain health and development, and open the possibility of designing drugs that exploit this transporter to treat brain disorders.
The study, published June 16 in Nature, used low-temperature electron microscopy (cryo-EM) to image the transporter protein (called MFSD2A), combined with functional analyses and advanced methods of computer modeling and simulations, to discover how it handles its fatty acid cargoes.
“We now understand much better how this transporter brings fatty acids across the blood-brain barrier, and we can start to think about designing drugs that get into the brain by mimicking how these fatty acids interact with the transporter,” said study co-senior author Dr. George Khelashvili, an assistant professor of physiology and biophysics and an assistant professor of computational biophysics in the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine at Weill Cornell Medicine.
The blood-brain barrier is a special lining in small blood vessels in the brain that prevents most of the molecules flowing through these vessels from seeping out into brain tissues. The barrier is thought to have evolved as a way of protecting neural tissues from infections, inflammation and environmental toxins.
A problem for the developers of psychiatric, neurological and brain-cancer drugs is that the blood-brain barrier also prevents most types of drug molecule, including antibody-based drugs, from penetrating the brain. A similar barrier exists in the retina, which is an extension of the brain, so that most drugs for retinal diseases must be delivered directly by injection.
Scientists therefore have been eager to understand the workings of transporters such as MFSD2A, which sit in the outer membranes of the vessel-lining endothelial cells that form the blood-brain barrier. There, they selectively pull molecules from the bloodstream across the barrier so that they can move into the brain and retina.
MFSD2A is important for brain and retinal health because it transports docosahexaenoic acid (DHA), an omega-3 fatty acid that is needed to build, among other things, the membranes of brain and retinal cells. DHA cannot be produced in these tissues and generally comes from dietary sources, especially fish.
A team led by Dr. David Silver, a professor at Duke-NUS Medical School, had identified MFSD2A as the transporter for DHA in a 2014 study. Subsequent studies have linked disruptions of the gene for MFSD2A in humans and mice to severe neurological problems including microcephaly, the development of an abnormally small brain.
In the new study, Dr. Silver and his group teamed up with the laboratory of Dr. Filippo Mancia, a structural biologist at Columbia University and co-senior author of the study, and Dr. Khelashvili, a computational biophysicist. To image MFSD2A with cryo-EM and learn its structure they used the most stable version of this large and flimsy protein they could find, one from a tropical, chicken-like bird called the red junglefowl, to obtain a detailed, near-atomic-scale “picture” of the protein’s structure.
“The blood-brain barrier excludes the uptake of approximately 98 percent of drugs, limiting the treatment of neurological diseases,” Dr. Silver said. “The structural information we revealed in our study can be exploited to better design neurotherapeutics that can be transported by MFSD2A.”
“It’s extremely exciting to be able to view a protein’s shape at such a resolution,” said Dr. Mancia, an associate professor of physiology and cellular biophysics at Columbia University Vagelos College of Physicians and Surgeons whose post-doctoral fellow, Dr. Rosemary Cater, performed the structure determination. “We are talking about measurements of less than a billionth of a meter in size—and this information is critical to understand how it works at a molecular level.”
Dr. Khelashvili used large-scale atomistic ensemble molecular dynamics (MD) simulations, followed by detailed analysis of the MD data with advanced computational biophysics methods (such as Markov State Modeling) to “un-freeze” the cryo-EM structure and to study a dynamic molecular mechanism of how MFSD2A works. These computational approaches allow dynamic sampling of atoms in a protein over time, based on the laws of physics, and enable quantitative descriptions of the kinetics of dynamic processes.
Combined with the functional data from the team’s mutagenesis experiments on the MFDS2A gene —essentially altering specific amino acids in the protein sequence and observing the effects on its functions—the simulations and computational analyses revealed a detailed model of the mechanism by which this atypical MFS transporter mediates uptake of single-chain phospholipids into the brain.
The model indicates that binding of a sodium ion primes MFSD2A to accept one of the fatty acids it recognizes. Once the transporter engages with the ion and the fatty acid, it swivels the two major halves of its structure to bring its important cargo from the outside of a vessel-lining endothelial cell to the inside. This allows the fatty acid to make its way into the surrounding tissue.
The researchers view the development of their model as a big step toward designing drugs that can exploit MFSD2A to get into the brain or retina. For now, though, they plan to continue their mechanistic studies of MFSD2A to elucidate the initial steps in its transport cycle, and to determine why it transports some fatty acids but not others.