Researchers Explore the Microbiome, Medicine's Newest Frontier
By Amy Crawford
Portraits by John Abbott
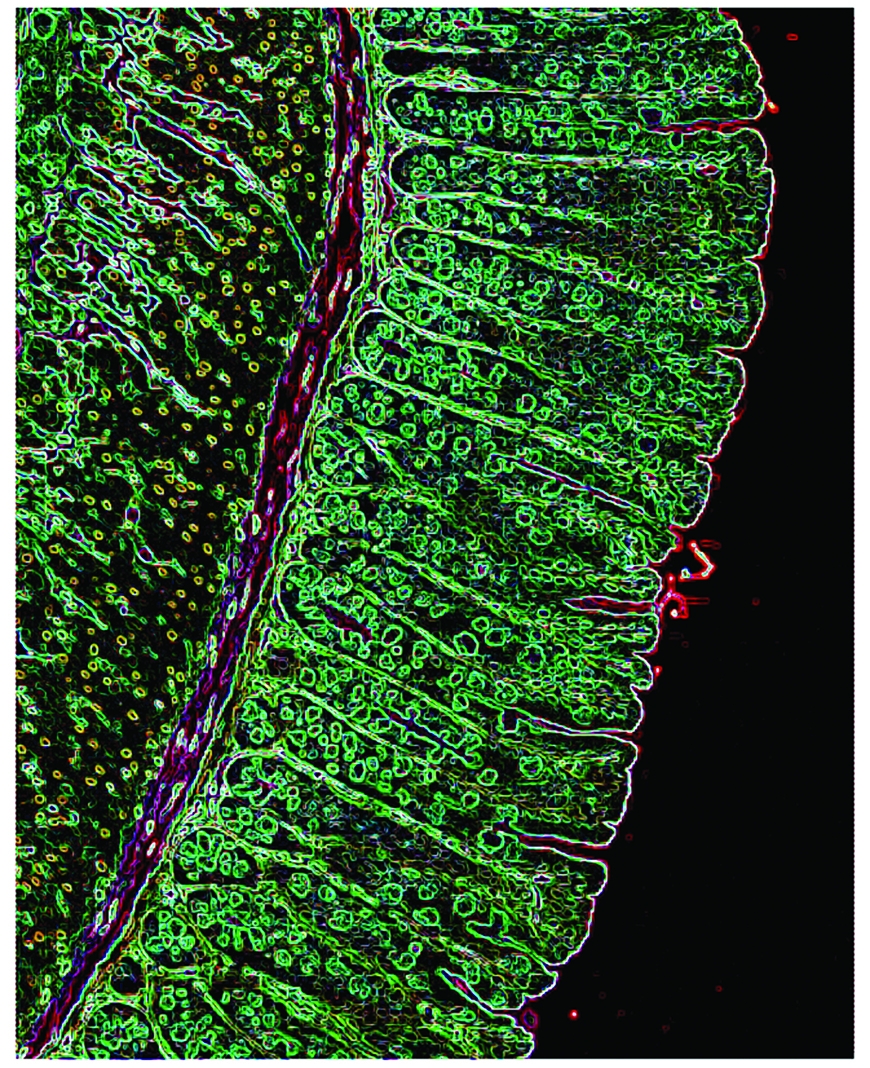
A color-enhanced image of the intestinal epithelium that lines the gastro-intestinal tract of a mouse. Scientific images: Drs. Gregory Sonnenberg and David Artis
The most exciting discoveries can begin with the humblest material — and for researchers in the lab of immunologist Dr. David Artis, that often means mouse droppings. Once an obliging rodent provides a stool sample, a technician uses a chemical buffer to break down bacterial cell walls, unleashing the coils of DNA within. After further chemical preparation to enrich the genetic information, the sample is fed into an Illumina MiSeq, a desktop machine half the size of an office photocopier that, over the course of about 48 hours, sequences billions of base pairs to reveal a catalog of hundreds of types of bacteria: the mouse's microbiome. "After painstaking collection and preparation, you load your samples and allow the machine to run overnight," explains post-doctoral researcher Dr. Lisa Osborne, who has analyzed her share of murine fecal bacteria in the name of better understanding how the microbiome works in humans. "A lot of sophisticated magic happens inside that box."
The technology may not look flashy, but it's enabling a revolution in how scientists think about the bacteria that share our bodies — no longer as mere pathogens, but as members of a tiny ecosystem that coevolved with us, and on which our health depends. Dr. Artis and his colleagues hope these communities of microbes can offer insight into one of the most confounding problems in modern medicine: the set of painful chronic conditions known as inflammatory bowel disease (IBD). But IBD is not the only reason scientists at Weill Cornell and elsewhere are increasingly interested in microbiota. It turns out that the human microbiome may have far-reaching impact throughout the body, influencing how our immune systems develop, how our food is metabolized — and even, perhaps, the peculiarities of our personalities. New knowledge about the complex web of relationships between humans and the microbes that live within us is calling into question not only our understanding of disease, but of what it means to be human. "This is one of the major topics in contemporary biomedicine, and it's profoundly reshaping the way we think about health and disease and individuality," says Dr. Carl Nathan, the R. A. Rees Pritchett Professor of Microbiology and chairman of microbiology and immunology. "I grew up thinking that a given person has one genome, one set of genes that you inherit from your two parents. That's much too simple."
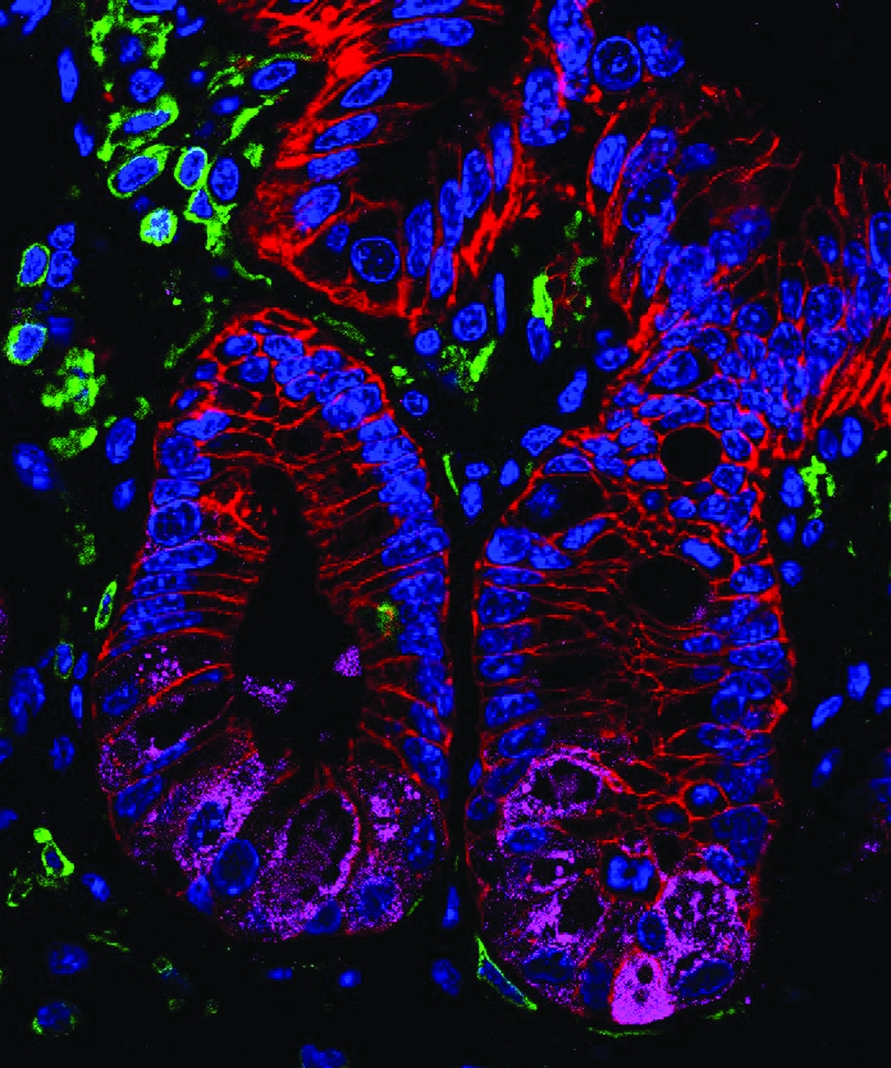
A stained histologic section of intestinal tissue isolated from healthy mouse.
Medical training taught Dr. Nathan that the hereditary genome he learned about in school was not the only one that makes us who and what we are. There's also the somatic genome — the accumulated mutations and re-arrangements that some cells undergo, either as part of an abnormal process that can lead to cancer or as part of a healthy immune system, which adapts to recognize the myriad pathogens a person encounters throughout life. Another code is found within mitochondria, organelles that power our cells and, scientists believe, evolved in multicellular organisms like humans from symbiotic bacteria. "Then there's the fourth genome," Dr. Nathan says. "That's the collective complement of genes of all the bacteria that normally reside in us. And the ways that this impacts medicine are almost countless."
In a suite of gleaming new labs on the fifth floor of the Belfer Research Building, some 20 people are working to unravel the mysteries of the microbiome. Some are hunched over laptops, while others collect data from a machine called a flow cytometer set up in a corner. In a culture room, several sit at biosafety cabinets, manipulating cells collected from mice or human patients.
The team is led by Dr. Artis, who was recruited as the Michael Kors Professor in Immunology. Widely considered a world leader in his field, he has recently been involved in studies that uncovered links between the immune system's response to gut bacteria and systemic allergies, as well as how the immune system keeps gut bacteria where they belong. Last year, Dr. Artis, along with some of his longtime collaborators, was lured away from the University of Pennsylvania to head Weill Cornell's new Jill Roberts Institute for Research in Inflammatory Bowel Disease.
When Dr. Artis, who grew up in Scotland, was an undergraduate in the early '90s, he took a course on evolution and became fascinated by how the mammalian immune system had evolved in conjunction with the pathogens that infect us. After earning a Ph.D. in immunology at the University of Manchester, he crossed the Atlantic to do postdoctoral work at Penn, where he would later join the faculty. Some of his early research there centered on the immune system's interaction with helminths, tiny worms that can make their home in human intestines, into which they find their way via under-cooked meat or contaminated water. Broader interest in the human microbiome as anything but pathogenic had yet to take hold, but Dr. Artis and other researchers were beginning to recognize something that ran counter to our previous understanding of these parasites as little more than uninvited passengers that make us sick. "We were interested in the pathogens that infect us, and one class of pathogens is worms," Dr. Artis explains. "The interesting thing is that to eradicate worms, the body mounts the Type 2 inflammatory response. It's the same type of response in allergies, only there it's reacting to innocuous antigens in peanuts and milk products and so forth."
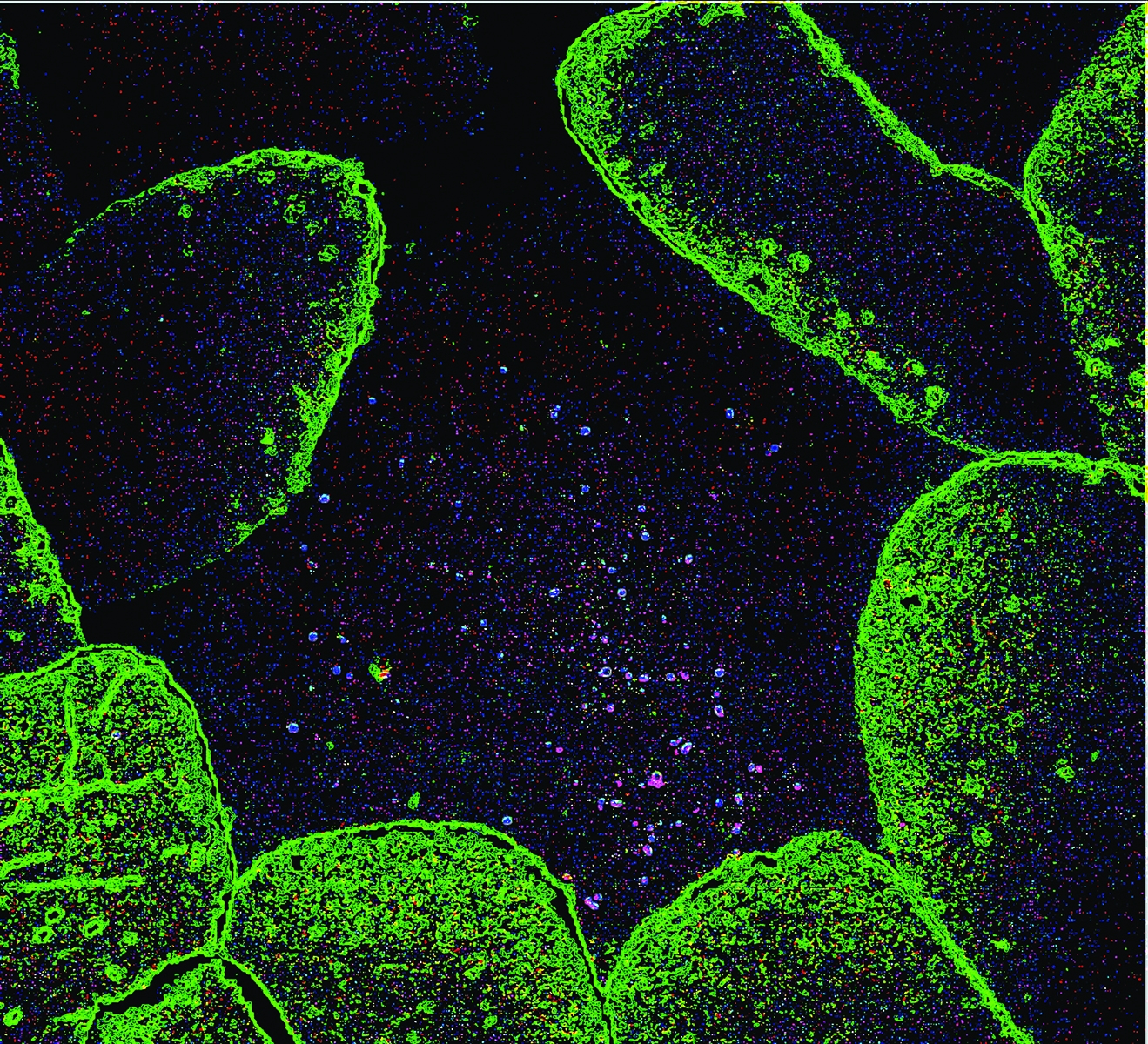
A color-enhanced tissue section from a healthy mouse showing the presence of normal microflora.
Over most of human history, the Type 2 response was an effective way to combat common parasites, and it's still called into action in much of the world, where helminths remain a problem. But in the United States and other industrialized countries, sanitation and medicine have virtually eliminated intestinal worms, leaving the Type 2 response a weapon without a proper target. That mismatch may contribute to the startling increase in allergic disease, asthma and other immune disorders, including certain forms of IBD.
As Dr. Artis' research looked at the ways in which immunity in the presence or absence of parasites could be involved with allergic reactions and chronic inflammation, a great shift was taking place in how researchers, doctors and even the general public think about other organisms that live in our bodies. It was a shift that paralleled the discovery of microorganisms themselves in the late 17th century, when the Dutch scientist Antonie van Leeuwenhoek trained his homemade microscopes on droplets of rainwater. "Like most discoveries in science," Dr. Artis says, "these quantum leaps are triggered by new technologies that allow us to see differently." A decade ago, studying the organisms living in someone's colon would have required culturing them in a petri dish, a time-consuming technique that could only begin to reveal the multitudes of bacteria that make up a complete human microbiome. That has changed, largely thanks to an international science project that some have compared to the 1969 moon landing in both its historical importance and its legacy of innovation.
In 2000, President Bill Clinton and British Prime Minister Tony Blair appeared on television to announce that an international team of scientists had completed a rough draft of the human genome, some 3 billion base pairs that make up roughly 20,500 genes. The project had been a massive undertaking, involving researchers in six countries working for more than a decade. In addition to the invaluable information about our own DNA that the project provided, it also spurred the development of new technology that would enable further discoveries. Today, commercially available genetic sequencing platforms like the Illumina MiSeq are considered de rigueur for any well-stocked research institution — Weill Cornell's Genomics Resources Core Facility has several — and what took the Human Genome Project years to accomplish can be done overnight. Now, researchers are using that technology to read and understand that fourth human genome, that of the bacteria that make their homes in our bodies. "Sequencing technology allows us to identify the microbiota at a level that we would never have been able to understand before," Dr. Artis says. "That technology has really accelerated our ability to profile the organisms in this complex ecosystem, and it also allows us to report how their composition changes in the context of disease."
Much as the sequencing of the human genome inspired the popular imagination a decade ago, today studies of the human microbiome have filtered from scientific journals and into the popular press. Breathless newspaper articles have told us how gut bacteria influence the workings of the mind, and that they might determine why some of us get fat while others stay slim on the same diet. In a 2013 New York Times Magazine cover story, the writer Michael Pollan recounted how sequencing the genes of the 100 trillion bacteria in his own body led him to think of himself "in the first-person plural — as a superorganism, that is, rather than a plain old individual human being."
Dr. Greg Sonnenberg, an assistant professor of microbiology and immunology in medicine, sees this as an asset to science. "The current level of excitement is fantastic," says Dr. Sonnenberg, who has collaborated on seminal studies with Dr. Artis and who was also recruited to lead a lab at the Roberts Institute. "The more you learn about the microbiome, the more it just touches upon everything; it is involved in probably every human disease out there. It's like the rainforest, where you can go through and find different bugs that may have the ability to provide therapeutic benefit in many diseases. And that's where the field is today. Now we need to get down to the nitty gritty in determining which species are important, which species are doing what and how are they interacting with each other. It's an extremely complex system."
Dr. Sonnenberg cautions that many recent papers based on sequencing data have likely uncovered mere correlations. While some members of the microbiome are clearly associated with certain medical conditions, he explains, that doesn't mean the bacteria caused the conditions. Much more work must be done to understand the functions of the bacteria that make up the human microbiome, and how the chemical signals and byproducts they produce affect us and each other. "Hopefully," he says, "that's going to translate to more research being done that advances us to the point where it will benefit patients more directly."
There is already one way in which doctors are using knowledge of the microbiome to benefit patients. Clostridium difficile (C. diff.) is a highly antibiotic-resistant bacterium that causes severe diarrhea and kills some 14,000 Americans each year. Patients most at risk are those in whom antibiotics have wiped out beneficial gut bacteria, leaving the coast clear for C. diff. to grow unimpeded. So far the most successful treatment involves replacing those good bugs with a fecal transplant — that is, inserting the stool of a healthy volunteer into the colon of a C. diff. sufferer — and restoring the normal, healthy balance of gut bacteria. "The concept is both intriguing and somewhat repulsive," admits Dr. Charlie Buffie, who will finish his medical degree at Weill Cornell in 2016, having completed his doctoral work at Sloan Kettering through the Tri-Institutional M.D.-Ph.D. program. "But the efficacy of a fecal transplant has been strikingly high under the right circumstances." Fecal transplant cures about 90 percent of C. diff. patients, but doctors aren't sure exactly why. And because the treatment by its very nature is impossible to standardize, government regulators are uneasy and doctors are reluctant to use it in immunocompromised patients. Dr. Buffie, however, may have found a partial answer to those quandaries.

Dr. Charlie Buffie
The components of a fecal transplant are as numerous as those of the human microbiome itself. But one that seems to be especially effective in controlling a C. diff. infection is a related species called Clostridium scindens. In a study published last year in Nature, Dr. Buffie and colleagues in the Sloan Kettering lab of immunologist Dr. Eric Pamer used C. scindens to defeat C. diff. in mice whose normal microbiomes had been disrupted with antibiotics. In the future, Dr. Buffie says, patients with C. diff. might be given precisely calibrated mixtures of beneficial bacteria or drugs that mimic the metabolic products of C. scindens that seem to prevent C. diff. from propagating. That would allow patients to avoid the potential safety risks — not to mention the ick factor — of a fecal transplant. Says Dr. Buffie: "Being able to isolate, define and construct compositions of bacteria that we know have positive effects and that do not have negative effects — that's definitely an attractive solution."
This line of research also holds promise for IBD patients, says Dr. Randy Longman '07, an assistant professor of medicine in gastroenterology and alumnus of the Tri-Institutional M.D.-Ph.D. Program who joined the Jill Roberts Center for Inflammatory Bowel Disease in 2013. Preliminary evidence suggests that fecal transplantation could help those with a form of IBD called ulcerative colitis, and Dr. Longman and his colleagues are working to figure out why. "The idea is to be able to get specific about the microbes," he says. "If we isolate some of these bugs from patient samples and then put them into gnotobiotic mice we may be able to understand how these microbes interact with the immune system within the intestine."
Although Dr. Longman's primary occupation is research, he spends one day a week in clinical practice, working to help patients manage their illness. "Inflammatory bowel disease, epidemiologically, affects people in the prime of life," Dr. Longman notes. "So many of the patients that I'm seeing for initial diagnoses are young people with so much of their lives in front of them. There is a tremendous need for new medicines and new therapeutic strategies." Like his colleagues, Dr. Longman is optimistic that cracking the secrets of the microbiome will lead to better treatments, and his patients will one day get relief from the stress of living with a chronic condition. "Right now, treating IBD is a management thing," he says. "We don't cure it right now. But we do hope for that."
Today, fecal transplantation remains the best microbiome-based treatment available. But as research points to gut bacteria's involvement with a variety of other ailments, scientists are hoping that future patients could be helped by targeted probiotics or drugs modeled after the chemical signaling of good bacteria. "There may be a point where it isn't necessary to cultivate certain bacteria in your body," Dr. Nathan says, "but rather to take a pill that provides the compounds those bacteria are making — to do the job they do, but in a more orderly, defined, predictable, consistent, safe way." In the future, such treatments might be used not only for C. diff. and IBD, but eventually for metabolic disorders, obesity and even neurological problems. "Whether the food you eat influences a predisposition to atherosclerosis — that's controlled by the bacteria in the body," Dr. Nathan says. "There are influences on behavior, on weight gain, probably on asthma. There's a connection to autism that's recently been reported." The microbiome may have an impact on every system in the human body, he stresses, and its importance is inestimable.
Dr. Artis echoes Dr. Nathan's enthusiasm, and notes that most breakthroughs are yet to come. He draws an analogy to the years after van Leeuwenhoek's microscope first revealed the hidden world in a water droplet. "The pace of discovery is so rapid; this field has really exploded," he says. "But in terms of understanding the complexities of microbiota in the body, we're in our infancy."
Living Underground
The New York Subway System Hosts a Complex Bacterial Ecosystem, Too
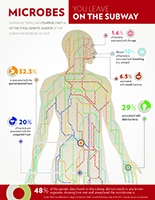
Just as each human body holds a complex ecosystem of bacteria in the gut, every major metropolis is home to a medley of bacteria and pathogens, coexisting with that city's residents. Until recently, little was known about these native microbial communities, which surround us in streets, buildings and public transit areas.
Using the subway system as their testing ground, Weill Cornell investigators fanned out beginning in June 2013 and collected samples of hundreds of DNA from bacterial, viral, fungal, and animal species — like insects and domestic pets — in the underbelly of New York City. They then compiled the data and turned it into an interactive pathogen map, dubbed PathoMap, and recently published their findings in Cell Systems.
While most of the collected microbes are harmless, some are not — including live, antibiotic- resistant bacteria, which were found in 27 percent of the samples. Two samples included DNA fragments of anthrax, and three carried a plasmid associated with Bubonic plague. Reassuringly, those five were discovered at very low levels and showed no evidence of being alive. Other DNA — half of what was collected, in fact — could not be identified as any known organism, because the databases against which they are compared are still incomplete.
While this might sound troubling, there's no need to worry right now, says the study's senior investigator, Dr. Christopher Mason, the WorldQuant Foundation Research Scholar and an associate professor in Weill Cornell's Department of Physiology and Biophysics and in the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Al-Saud Institute for Computational Biomedicine. These apparently virulent organisms are not linked to widespread sickness or disease in this environment, Dr. Mason says. "They are instead likely just the co-habitants of any shared urban infrastructure and city," he says, "but additional testing is needed to confirm this."
The knowledge that these bacteria are present and having no obvious negative effect on the 5.5 million daily subway riders demonstrates that most of them are neutral to human health, he adds. They may even be helpful, as they can out-compete dangerous bacteria. "The presence of these microbes and the lack of reported medical cases is truly a testament to our body's immune system," Dr. Mason says, "and our innate ability to continuously adapt to our environment." Would these pathogens be typical for other cities? With the aim of answering that question, collaborators are collecting samples from airports, taxis and public parks in 15 other cities around the world under a recent grant from the Sloan Foundation.
The PathoMap project involved investigators from Weill Cornell, five additional New York City medical centers and more than a dozen national and international institutions. Over the course of 17 months, medical students and other volunteers used nylon swabs to collect DNA from turnstiles, benches, railings, trashcans and kiosks in all operating subway stations across the five boroughs. The team also collected samples from inside trains, swabbing seats, doors, poles and handrails. They time-stamped each sample and tagged it using a GPS system, later sequencing about 1,500 samples (out of more than 4,200 collected) and analyzing those results. "PathoMap establishes the first baseline data for an entire city," Dr. Mason says, "revealing that 'molecular echoes' of commuters appear on all surfaces — from the bacteria on their skin to the food they eat, and even from the human DNA left behind, which matched U.S. Census data."
The data on New York City's ecosystem — an ingredient in building a smart city — already has potential real-world applications. Researchers could monitor the system for changes that would signal disease or a potential threat, or someday create a live model tracking real-time changes to this urban microbiome. The PathoMap, Dr. Mason says, is just the beginning.
— Anne Machalinski
This story first appeared in Weill Cornell Medicine, Vol. 14, No. 1.